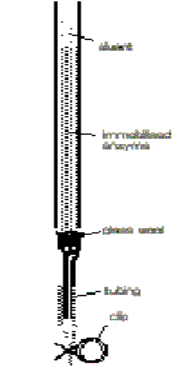
Protein and Enzyme Technology
Practical Exercise
Aim
To
investigate some of the properties of immobilised enzymes.
Introduction
The
four-session practical exercise is designed to give some insight into the
preparation and properties of immobilised enzymes.
It
consists of three parts:
- Preparation and properties of covalently-immobilised a-amylase.
- Preparation and properties of non-covalently-immobilised a-amylase.
- The use of a packed-bed and stirred tank reactor.
Before
starting work, read through the Methods and Results sections. This
practical is very demanding and must be approached with thought and care. It
will be necessary to retain samples of the soluble enzyme before and
after coupling (as well as the immobilised enzyme, of course) for protein and
activity assay (see Appendix A) in
order to determine the amount of enzyme coupled. You are expected to work in
teams of three with a named team leader. You must plan and organise your
experiments carefully, for which marks will be awarded. a-amylase
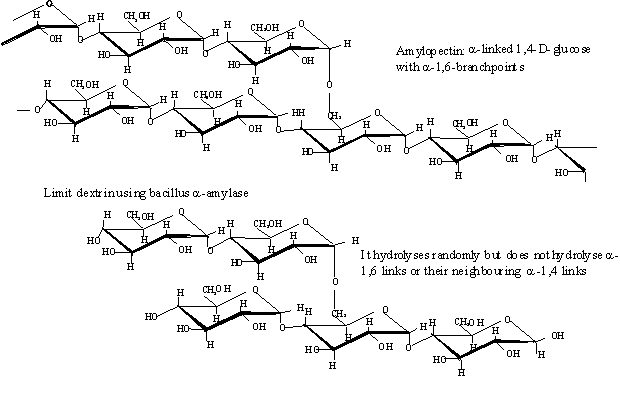
is an enzyme produced and purified from Bacillus. It hydrolyses the a-1-4 links
in starch randomly along its structure (i.e. it is an endo-glycosidase). It
cannot hydrolyse a-1-6 links. Complete hydrolysis of starch by a-amylase
produces a mixture of short glucose oligomers (e.g. maltose, maltotriose), some
limit dextrin containing a-1-6 links but relatively little glucose. The quality
of hydrolysed starches is given in terms of its dextrose equivalent (DE),
which equals the percentage of the starch that is hydrolysed. ('Dextrose' is
another word for glucose).
a-Amylase
is assayed by the creation of new reducing (terminal; equivalent in reducing
power to glucose) sugar by the catalysed hydrolysis of soluble starch.
In
this practical, a-amylase is immobilised by means of covalent and
non-covalent binding to solid supports. The amount of enzyme attached to the
supports is determined and the activities of the immobilised enzymes are
compared to that of the free (non-immobilised) enzyme. Packed bed reactors
containing the immobilised enzymes are prepared and their ability to hydrolyse
starch compared.
Plan
During week 1 you should
- Prepare covalently-immobilised a-amylase
- Prepare non-covalently-immobilised a-amylase
- Construct standard curves for protein and reducing sugar (see Appendix A
for details). Draw these before week 2 when they will be required.
During
week 2 you should
- Wash the covalently-immobilised a-amylase
- Wash the non-covalently-immobilised a-amylase
- Assay samples from the preparations; see 'Assays' later
During
weeks 3 and 4 you should run the stirred tank and packed bed reactors and
analyse their products.
Covalent
Immobilisation of a-amylase (Methods
Enzymol. 44, pp. 98-99)
|
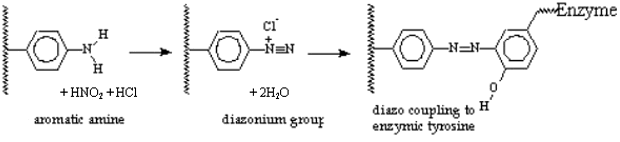
Safety note: The following makes use of nitrous acid.
This gives off toxic brown fumes of nitrogen oxides, if warmed. It is
important from both a safety and experimental point of view that it is kept ICE-COLD. |
Week
1. Weigh out 1.0 g Enzacryl AA gel. (This is a gel based on a
polyacrylamide matrix with aromatic amino groups present as the reactive
moieties). Add to 50 ml ICE-COLD 2 M HCl, stir at 0°C, and gradually add
40 ml ICE-COLD 2% sodium nitrite solution (Together these produce nitrous acid,
HNO2).
NaNO2 + HCl ![]() HNO2
+ NaCl
HNO2
+ NaCl
Keep
the beaker surrounded by ice during addition. This should take about 10 - 15
minutes. (This creates the reactive diazonium groups from the aromatic amino
groups. If these are allowed to warm up, they decompose to give nitrogen gas
with the loss of their specific reactivity)
Stir
for another 15 minutes, then filter the gel on a filter paper disc on a Buchner
funnel, by suction. Wash with 200 ml ICE-COLD 20 mM phosphate buffer, pH
7.0, 0.1 mM CaCl2 (a-amylase buffer). Add the buffer in 25 ml batches,
and KEEP IT COLD. At this stage, the amino groups of the gel matrix should be
diazotized. Quickly scrape gel off the filter into a test tube. Add 5 ml of an ICE-COLD
solution of a-amylase (2 mg/ml in phosphate buffer). Cap, label ('Cov')
swirl in an ice bath for about an hour and leave in the fridge until next week.
(This allows the diazo groups to covalently couple to the tyrosine phenolic
groups on the enzyme)
Non-Covalent
Immobilisation of a-amylase
|
Safety note: The
following makes use of a fine powder. Treat it with care and do not allow this
to form a dust cloud Clean all spillages with slightly damp tissue. |
Resin
structure
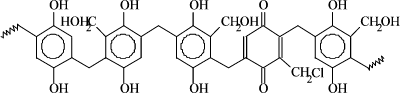
Week
1. Weigh out 0.6 g dry phenolic resin (invented and patented
by M. F.
Chaplin, J Chem Soc., Perkin 1, 1979, pp 2144-2153), suspend in 20 ml 20 mM
K phosphate pH 7.0, 0.1 mM CaCl2 (a-amylase buffer) for 10 minutes.
Filter and re-suspend in 5 ml of 2 mg/ml a-amylase in the 20 mM phosphate
buffer. Cap, label ('Non') swirl for 30 min and leave in the fridge
until next week
N.B.: Keep a
solution of the free enzyme (0.5 ml 2 mg/ml) similarly capped in the fridge as
a comparison for both 'Cov' and 'Non' above (label 'Enz').
Covalent Immobilisation of a-amylase
Filter
the gel ('Cov'), using a fluted filter paper, into a test tube; use 5 ml
of 20 mM K phosphate buffer pH 7.0 to aid this process. Keep about 2 ml of the
filtrate for assay of unbound protein and activity, (Label it 'Cov-supernatant')
|
NB. As you added 5 ml of buffer to the 5 ml
of original enzyme solution, any enzyme remaining in solution has been
diluted by a factor of two. |
Wash
gels to remove any free enzyme, using 3 x 20 ml batches of 20 mM potassium
phosphate/500 mM NaCl pH 7.0 ('high salt buffer'). Let the gel damp-dry briefly
between each 20 ml portion of buffer. Wash once more in the same buffer without
NaCl, and re-suspend the gel in 5 ml in 20 mM K phosphate (pH 7.0). Label it 'Cov-immobilised'
and refrigerate until next week.
Non-Covalent Immobilisation of a-amylase
Filter
the gel ('Non'), using a fluted filter paper, into a test tube; use a
further 5 ml of 20 mM K phosphate buffer pH 7.0 to aid this process. Keep about
2 ml of the filtrate for assay of unbound protein and activity, (Label it 'Non-supernatant'),
.
|
NB. As you added 5 ml of buffer to the 5 ml
of original enzyme solution, any enzyme remaining in solution has been
diluted by a factor of two. |
Wash
gels to remove any free enzyme, using 3 x 20 ml batches of 20 mM K phosphate
buffer pH 7.0. Re-suspend in 5 ml of this buffer. Label it 'Non-immobilised'
and refrigerate until next week.
At
this stage During Week 2 you should have the following samples:
- Cov-supernatant:
unbound a-amylase; left over from covalent
immobilisation (2 ml of not more than 1.0 mg/ml)
- Cov-immobilised:
covalently bound a-amylase; 1 g dry gel containing not
more than 10 mg enzyme.
- Non-supernatant:
unbound a-amylase; left over from non-covalent
immobilisation (2 ml, not more than 1.0 mg/ml).
- Non-immobilised:
non-covalently bound a-amylase; 0.6 g dry gel containing not
more than 10 mg enzyme.
- Enz: stored
free a-amylase (0.5 ml of 2.0 mg/ml). Some of
this should be refrigerated for next week.
Assays (See Appendix for
details of the procedures)
|
Note: you have to dilute the
samples until they contain about 50 mg/ml protein so that your determinations
are in the right range for the assays. Do not forget to allow for these
dilutions when you determine the protein content and a-amylase
activity of the original. |
Dilute
samples 'cov-supernatant' and 'non-supernatant' 1:20 v/v and
sample 'enz' 1:40 v/v. Assay these diluted samples of 'cov-supernatant',
'non-supernatant' and 'enz' for protein content (Assay 1) and a-amylase
activity by production of reducing equivalents (Assay 3).
Ensure that you record how you dilute these samples in your notebook.
Although
you know the concentration of protein in sample 'enz' (2 mg/ml), you
will probably get a different value as determined in the Dye-binding assays due
to the different standard protein (bovine serum albumin not a-amylase)
used. Use this value to 'correct' the protein concentrations (i.e. if the
apparent Dye-binding concentration of sample 'enz' is 1.5 mg/ml then all
final protein concentrations as determined by the Dye-binding method should be
multiplied by the factor 2.0/ 1.5).
Tabulate
the protein concentration (mg/ml, uncorrected and corrected) and activity (mmol
reducing sugar released/min/ml and mmol reducing sugar
released/min/mg) of the diluted and original undiluted samples cov-supernatant,
non-supernatant and enz. By allowing for the volumes of solutions
used in the binding (5 ml) and filtering (another 5 ml), tabulate also the
total protein content of the supernatants.
This
Table will allow you to calculate:
- the (corrected) weight of a-amylase protein not bound to each
immobilisation matrix from the protein concentrations in the unbound
residual enzyme samples cov-supernatant and non-supernatant.
Make sure that you allow for the dilutions and the final volume of the
wash solution (10 ml) and the correction for the use of the bovine serum
albumin standard.
- the weight of protein bound to each immobilisation matrix, by
subtracting the (corrected weight of) protein not bound (from above) from
the amount added (5 ml x 2 mg/ml = 10 mg).
- the percentage of the enzyme protein that was added that is
bound to each immobilisation matrix.
- the specific activity of the original a-amylase
solution used (sample 'enz'); Note that the specific activity
equals the activity of one mg a-amylase protein. the units are in mmol
reducing sugar released per min per mg of protein).
- the specific activity of the unbound a-amylase
solutions left after each of the immobilisation processes. Note that these
would be expected to be identical to the specific activity of the original
a-amylase solution used (sample 'enz') unless some
denaturation occurred in the immobilisation process.
Note: you have
to dilute the samples until they contain about 50 mg/ml
protein so that your determinations are in the right range for the assays. Do
not forget to allow for these dilutions when you determine the protein content
and a-amylase activity of the original solutions.
Figure 1
Prepare
two small packed bed reactors containing all of the covalently and
non-covalently immobilised a-amylase ('cov-immobilised' and 'non-immobilised').
Do not allow them to run dry. (Note that if they are allowed to develop an air
lock, they will not flow and must be repacked) Run 5 ml of 1% starch in 20 mM K
phosphate pH 7.0, 0.1 mM CaCl2. through each column.
|
|
Reduce the flow
rate through the columns to about 1 ml per 10 min (i.e. about one drop every
30 seconds), allow the starch solution to run through for about 15 min and
then collect 2 ml from each column for analysis, label 'cov-eluent'
from the 'cov-immobilised' column and 'non-eluent 'from the
'non-immobilised' column and set to one side. Wash
the packed bed reactors with 3 column volumes of phosphate buffer (20 mM K
phosphate, pH 7) without starch and store refrigerated until week 4. Assay
the partially-hydrolysed starch samples cov-eluent and non-eluent
for reducing equivalents (Assay 2). |
Remove the
gels ('cov-immobilised' and 'non-immobilised') from the columns.
Using all of the samples of immobilised enzymes determine (separately) their
activity in a stirred reactors (beaker) containing 50 ml of 1.0% w/v starch in
20 mM K phosphate pH 7.0, 0.1 mM CaCl2. Withdraw samples at intervals (e.g. 1
min, 5 min, 10 min, etc.) to determine their reducing sugar content.
From
the assay of the stirred tank and packed bed reactors you should calculate :
- the concentration of reducing equivalent in the
partially-hydrolysed starch samples (mmoles of reducing equivalent produced
per ml per reactor).
- the productivity of the reactors (mmoles of reducing
equivalent produced per minute per reactor),
- the fractional conversions, X (X = moles of reducing
equivalents produced/moles of potential glucose units in the starch
solution); Note that to calculate the number of moles of potential glucose
in the 1% starch, solution, the apparent M.Wt of potential glucose is 180
- 18 = 162, as water is necessary to release the glucose; the complete
hydrolysis of 162 g of starch produces one mole (180 g) of glucose, Also
remember that the number of moles in a sample is the weight in grams
divided by the weight of one mole (i.e. weight/ M.Wt). Also, note that because
a-amylase cannot hydrolyse limit dextrins, maltose, maltotriose or
maltotetraose, the highest value expected for the fractional conversion,
X, is about 0.2.
- the dextrose equivalent DE of the products (in this case the
fractional conversion, X, expressed as a percentage),
- the activity of the immobilised enzymes (mmoles
of reducing equivalent produced per minute per g resin).
- the specific activity of the immobilised enzymes (mmoles
of reducing equivalent produced per minute per mg enzyme) by using the
known amount of protein immobilised (determined previously).
- the effectiveness factors for the immobilised enzymes. Note that
the effectiveness factor is the specific activity of the immobilised
enzyme divided by the specific activity of an equal quantity of the free
enzyme (calculated previously).
Assay
samples 'cov-immobilised', 'non-immobilised' and 'enz' for
a-amylase activity by loss of iodine reactive material (Assay 4).
This reaction may be very rapid with excess free enzyme. For the free and
immobilised enzymes estimate the % digestion of the starch when the iodine
reactive material has been used up, by comparing these results with your
specific activity results from the production of reducing equivalents.
- determine the relative specific activities of the immobilised
enzymes compared with the free a-amylase. Use the reciprocals of the
times needed to decolourise the blue starch-iodide divided by the amounts
of a-amylase protein present.
- compare these specific activities with those calculated earlier.
Explain your results on the basis that starch molecules, once next to an
immobilised a-amylase, have difficulty diffusing
away due to their bulk. Thus, immobilised a-amylase is
expected to produce some starch that is completely hydrolysed before other
starch molecules are hydrolysed at all, whereas free a-amylase
hydrolyses all starch molecules roughly equally.
Practical:
Appendices
Assays
Note that all assays should be done in duplicate, where possible. Ensure
all the cuvettes are clean by checking their absorption against each other at
the assay wavelength before use.
1
Dye-binding Protein Assay
1.5 ml of protein sample solution (0 - 50 mg/ml) is mixed with 1.5 ml
Coomassie blue reagent (0.6% dye in dilute perchloric acid). Use 1.5 ml
distilled water plus 1.5 ml Coomassie blue reagent as blank to zero the
spectrophotometer. Read the absorbency at 620 nm.
A
standard curve is prepared by using the stock solution of bovine serum albumin
(BSA, 50 mg/ml) using at least four data points in duplicate.
e.g. 0.4 ml stock + 1.1 ml water (= 0.4/1.5 x 50 mg/ml =
13.3 mg/ml), 0.8 ml stock + 0.7 ml water, etc. N.B. only the
concentration within the 1.5 ml 'sample' solution is relevant; the (constant)
amount of reagent added is not relevant for sample concentration calculations
2
Reducing Sugar Assay
2.0 ml of DNS reagent (ready prepared; 3,5-dinitrosalicylic acid and sodium
potassium tartrate dissolved in dilute sodium hydroxide) is added to sample
(200 ml, 0.2 ml), containing 0 - 2 mg reducing sugar (i.e. 0 - 10
mg/ml). The tube is placed in a boiling water bath and the solution heated at
100°C for 5 minutes. Rapidly cool in ice to room temperature. Use 0.2 ml
distilled water plus 2.0 ml DNS reagent, heated as above, as blank to zero the
spectrophotometer. Read absorbency at 570 nm. A standard curve is prepared by
using the stock solution of maltose (10 mg/ml) using at least four data points
in duplicate. e.g. 0.05 ml stock + 0.15 ml water (= 0.05/0.2 x 10 mg/ml = 2.5
mg/ml), 0.1 ml stock + 0.1 ml water, etc. N.B. only the concentration within
the 0.2 ml 'sample' solution is relevant; the (constant) amount of reagent
added is not relevant for sample concentration calculations. You are reminded
that the M.Wt. of maltose is 342 and maltose contains a single reducing group
(i.e. 342 g maltose contains one mole of reducing group/equivalent). For your
graphs, you must calculate the molar concentration of reducing groups in the
standard maltose solutions.
3
Assay of a-amylase by production of reducing equivalents
Add 0.8 ml 20 mM K phosphate (a-amylase buffer) to 0.2 ml soluble enzyme in
phosphate buffer (containing about 10 mg amylase).
Note that the enzyme solutions must be diluted before they are assayed).
Pre-incubate for about 4 minutes at 37°C. Add 1.0 ml, 1% starch in phosphate
buffer (pre-warmed to 37°C). Incubate for exactly 5 minutes at 37°C. Stop the
reaction by removing 0.2 ml of the incubated mixture and adding this to 2 ml of
DNS reagent.
The
tube should be placed in a boiling water bath and the solution heated at
100°C for 5 minutes to develop the reducing sugar assay colour. Rapidly cool in
ice to room temperature and read absorbency at 570 nm. Use 0.1 ml buffer plus
0.1 ml starch plus 2.0 ml DNS reagent, heated as above, as a blank to zero the
spectrophotometer. Note that the reducing sugars in only 0.2 ml of the 2.0 ml
in the 37°C incubation mixture is used in the reducing sugar assay and
allowance should be made for this when calculating the amount of reducing sugar
produced by the enzyme in the 0.2 ml original sample.
4
Assay of a-amylase by loss of iodine reactive material
Make a mixture of 0.1 ml buffer plus 0.1 ml starch for use as blank. Add one
drop to one drop of K phosphate containing 0.05% iodine. A blue coloration will
be observed.
Free
enzyme assay: Add 0.8 ml K phosphate (20 mM, pH 7, 'a-amylase
buffer') to 0.2 ml enzyme (containing about 10 µg free amylase). Incubate for 4
minutes at 37°C. Add 1 ml 1% starch (pre-warmed to 37°C) and incubate at 37°C.
At known times (e.g. 0, 30 s, 1, 2, 5, 10 min etc), remove 1 drop and drop into
1 ml K phosphate containing 0.05% iodine.
Immobilised
enzyme assay: Add 1.0 ml K phosphate (20 mM, pH 7, 'a-amylase
buffer') to half the immobilised enzyme. Incubate for 4 minutes at 37°C as
above. Add 1 ml 1% starch. Keep the immobilised enzymes agitated. At known
times (e.g. 0, 30 s, 1, 2, 5, 10 min etc), remove one drop and drop into one
drop of K phosphate containing 0.05% iodine. In both assays, blue coloration
will be observed while macromolecular starch is still present. The enzyme
activity is inversely proportional to the time taken. If no blue colour is
observed in the first samples, repeat the assay as either (1) the reaction has
already occurred at to rapid a pace, or (2) you forgot to add the enzyme/
iodine/starch/etc.